Molecular Biomedicine | Random-PE: an efficient integration of random sequences into mammalian genome by prime editing

Open the phone and scan
Prime editing (PE) enables efficiently targeted introduction of multiple types of small-sized genetic change without requiring double-strand breaks or donor templates. Here we designed a simple strategy to introduce random DNA sequences into targeted genomic loci by prime editing, which we named random prime editing (Random-PE). In our strategy, the prime editing guide RNA (pegRNA) was engineered to harbor random sequences between the primer binding sequence (PBS) and homologous arm (HA) of the reverse transcriptase templates. With these pegRNAs, we achieved efficient targeted insertion or substitution of random sequences with different lengths, ranging from 5 to 10, in mammalian cells. Importantly, the diversity of inserted sequences is well preserved. By fine-tuning the design of random sequences, authorswere able to make simultaneously insertions or substitutions of random sequences in multiple sites, allowing in situ evolution of multiple positions in a given protein. Therefore, these results provide a framework for targeted integration of random sequences into genomes, which can be redirected for manifold applications, such as in situ protospacer adjacent motif (PAM) library construction, enhancer screening, and DNA barcoding.

Here authors designed a simple strategy, named Random-PE, based on prime editing to introduce random sequences into the target region of the mammalian genome. In the Random-PE strategy, the pegRNAs were designed to harbor random sequences in-between the PBS and HA of the reverse transcriptase templates. They showed that the Random-PE strategy achieved efficient targeted insertion or substitution of random sequences up to 10 base pairs (bps), a theoretical diversity of ~ 107. To the best of their knowledge, this is the first attempt of trying to introduce random sequences into mammalian cells using pooled pegRNA library. During the preparation of the manuscript, a similar strategy using plasmid pegRNA library for random prime editing was developed in plant showing that 3-bp random sequences were able to be integrated into rice genome. Together with the result in plant, these works provide a framework for targeted integration of random sequences into the genomes, which can be redirected for manifold applications, such as in situ PAM library construction, enhancer screening, and DNA barcoding.
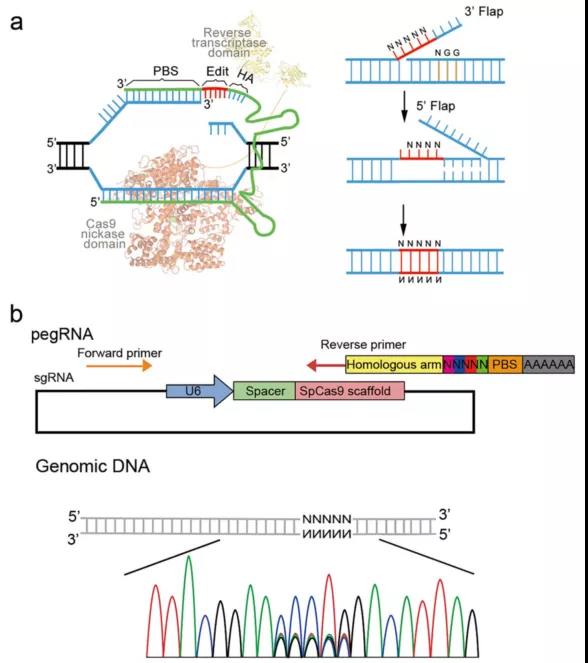
Design of Random-PE
Article Access: https://doi.org/10.1186/s43556-021-00057-w
Website for Molecular Biomedicine: https://www.springer.com/journal/43556
Looking forward to your contributions.


